
I recently looked at my personal blog and saw that I had an unpublished blog post there that contained a bunch of screenshots and had only one sentence incomplete sentence:
“like damn it. why did my gene have to be so awesome and conserved?????”
I thought it may be more relevant placing this entry here since it’s showing off how nerdy I was (and still am) about bioinformatics.
All little background information: this was last year in my third year of undergrad and I was taking an introductory course on computational biology, and this course had weekly computer labs and assignments associated with those labs. One of these assignments required us to look at a gene of interest (GOI), and use BLAST to see all the genes that pop up in the results. From there we were to get the sequences of those genes, align them using Seaview to your GOI and then produce a phylogenetic tree of all the genes.
My GOI of this assignment (and for a number of other assignments) was the CALM1 (calmodulin 1) gene. It’s one of three genes that encode for an identical calcium-binding protein that is a part of the subunits of phosphorylase kinase. After choosing this gene, I got its sequence and used NCBI’s BLAST and somehow got these results:

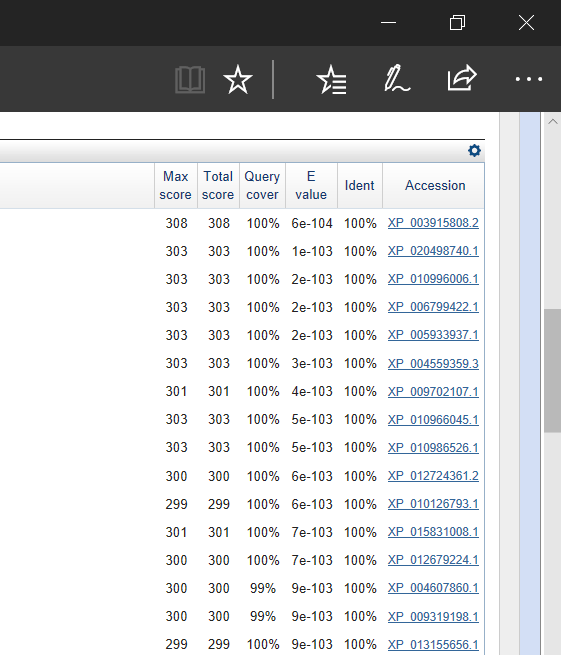
Now I don’t know about you, but getting more than 20 results with an E-value of 100% is pretty confusing, exciting and questionable. I remember feeling really skeptical but excited seeing such significant results, so I continued with the assignment and placed the top sequences into Seaview and got the following results:

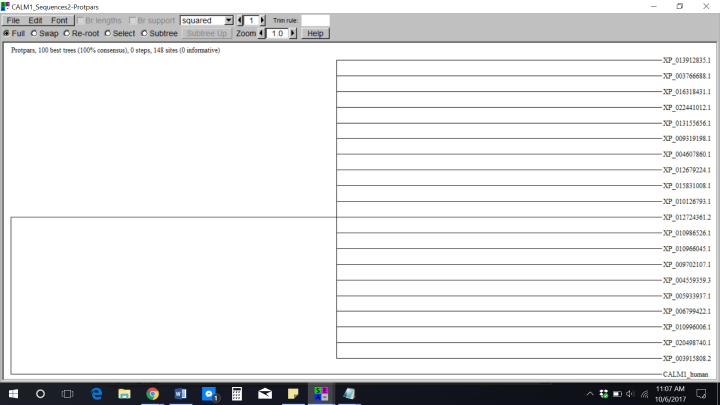
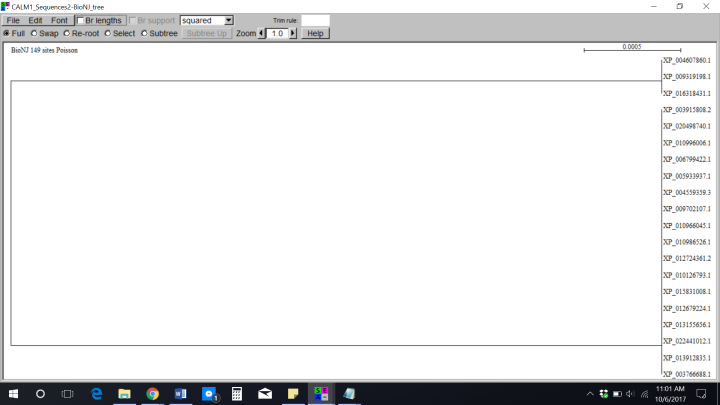
It was very odd seeing these results. I knew looking at them wasn’t going to give me the answers I needed to answer my assignment since I couldn’t really observe any differences in the trees. Even when I placed the sequences into Jalview gave me the same results as Seaview:
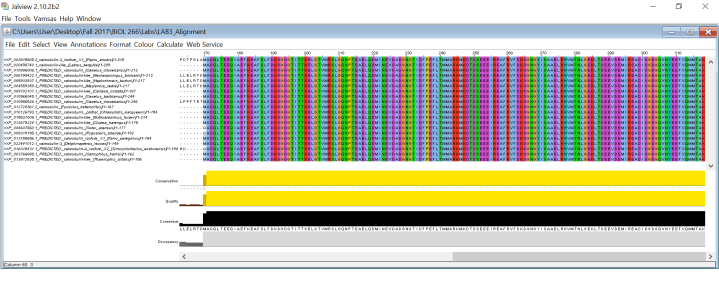
Now I’m not sure how I was able to get these results. When I tried doing this same assignment a couple of minutes ago, I didn’t get all of those genes with an E-value of 100%. Maybe I didn’t paste the whole sequence, maybe I used the wrong file, who knows. At the end of the lab, I showed my TA and
- He was shocked as I was when I got all those 100% E-values
- He suggested I choose another gene for this assignment
I was sad that I couldn’t use that gene, but it made sense since I couldn’t draw any conclusions on the homology of that gene.